Explain How Protein Structure Is Involved In Enzyme Specificity
Kalali
Mar 19, 2025 · 7 min read
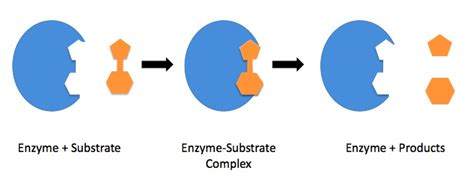
Table of Contents
Enzyme Specificity: The Crucial Role of Protein Structure
Enzymes are biological catalysts, accelerating the rate of virtually all chemical reactions within cells. Their remarkable efficiency and selectivity stem from a precise interplay between their three-dimensional structure and the molecules they interact with, known as substrates. Understanding how protein structure dictates enzyme specificity is fundamental to comprehending the intricacies of life's processes. This article delves into the complex relationship between protein structure and enzyme specificity, exploring the various levels of protein organization and their impact on substrate binding and catalysis.
The Four Levels of Protein Structure and their Contribution to Specificity
Enzyme specificity is not a random occurrence; it's a meticulously orchestrated consequence of the protein's intricate structure. Proteins, the workhorses of the cell, adopt a unique three-dimensional architecture through a hierarchical organization encompassing four levels of structure:
1. Primary Structure: The Amino Acid Sequence
The primary structure is the linear sequence of amino acids linked together by peptide bonds. This sequence, dictated by the genetic code, is the foundation upon which all higher levels of structure are built. The specific arrangement of amino acids, with their varying properties (size, charge, hydrophobicity, etc.), directly influences the folding pattern and ultimately, the enzyme's active site. The primary sequence determines the potential for forming specific secondary, tertiary, and quaternary structures. Even a single amino acid substitution can drastically alter an enzyme's activity and specificity. For instance, a change in a crucial amino acid within the active site could impair substrate binding or catalytic function.
2. Secondary Structure: Local Folding Patterns
The primary sequence spontaneously folds into regular, repeating structures known as secondary structures. The most common are α-helices and β-sheets. These structures are stabilized by hydrogen bonds between the backbone atoms of the polypeptide chain. The distribution and arrangement of these secondary structural elements contribute significantly to the overall three-dimensional shape of the enzyme and create the framework for the active site. The presence of specific secondary structural motifs can be crucial for substrate recognition and binding. For example, a particular loop region within a β-sheet might form crucial interactions with the substrate's functional groups.
3. Tertiary Structure: The Three-Dimensional Arrangement
The tertiary structure describes the overall three-dimensional arrangement of the polypeptide chain, encompassing all secondary structural elements. This structure is stabilized by a variety of interactions, including hydrogen bonds, ionic bonds, disulfide bridges (covalent bonds between cysteine residues), and hydrophobic interactions. The tertiary structure is critical for enzyme activity, as it determines the precise three-dimensional positioning of amino acid residues within the active site. The active site, a specific region within the enzyme's tertiary structure, is where the substrate binds and the catalytic reaction occurs. The intricate arrangement of amino acid side chains within the active site creates a unique microenvironment that is perfectly suited for substrate recognition and catalysis. Any disruption to the tertiary structure, such as denaturation by heat or pH changes, can lead to loss of enzyme activity.
4. Quaternary Structure: Multiple Polypeptide Chains
Some enzymes consist of multiple polypeptide chains, or subunits, each with its own tertiary structure. The arrangement of these subunits in space defines the quaternary structure. In multi-subunit enzymes, the interaction between subunits can be crucial for enzyme function and specificity. The interfaces between subunits often contribute to the formation of the active site or allosteric regulatory sites. Allosteric regulation involves the binding of a molecule at a site other than the active site, inducing conformational changes that affect substrate binding and catalytic activity. The quaternary structure plays a vital role in regulating enzyme activity and achieving the required specificity.
Mechanisms of Enzyme Specificity: Lock and Key vs. Induced Fit
Two primary models explain how enzyme specificity arises from protein structure: the lock-and-key model and the induced-fit model.
The Lock-and-Key Model
This classic model envisions the enzyme's active site as a rigid, pre-formed structure (the "lock") that precisely complements the shape of the substrate (the "key"). Only substrates with the perfect shape can fit into the active site, leading to a high degree of specificity. While this model provides a simplified explanation, it's now understood to be an oversimplification. Many enzymes exhibit flexibility, and their active site conformation can change upon substrate binding.
The Induced-Fit Model
The induced-fit model provides a more realistic representation of enzyme-substrate interactions. In this model, the enzyme's active site is flexible and undergoes conformational changes upon substrate binding. The substrate's binding induces a conformational change in the enzyme, optimizing the active site for catalysis. This dynamic interaction ensures a tight fit and enhances specificity. The conformational change can involve rearrangements of amino acid side chains, creating optimal interactions with the substrate and promoting catalysis. The induced-fit model better explains the ability of enzymes to accommodate structurally diverse substrates, provided they share essential functional groups.
The Role of Non-Covalent Interactions in Substrate Binding
The binding of the substrate to the enzyme's active site is primarily mediated by weak, non-covalent interactions, including:
- Hydrogen bonds: These form between polar groups on the substrate and the enzyme's active site.
- Ionic bonds (electrostatic interactions): These arise between oppositely charged groups.
- Hydrophobic interactions: These interactions occur between nonpolar groups on the substrate and the enzyme, often involving burying hydrophobic residues within the active site.
- Van der Waals forces: These are weak attractive forces that arise between atoms in close proximity.
The cumulative effect of these weak interactions contributes significantly to the overall binding affinity and specificity. The precise arrangement of these interactions within the active site determines which substrates can bind effectively and which are excluded. The strength and number of these interactions directly impact the enzyme's specificity and catalytic efficiency.
Factors Affecting Enzyme Specificity
Several factors beyond the basic protein structure influence enzyme specificity:
- Active site architecture: The three-dimensional arrangement of amino acid residues within the active site plays a paramount role in determining substrate specificity. The size, shape, and chemical properties of the active site dictates which substrates can fit and interact effectively.
- Substrate-enzyme interactions: The precise nature of the non-covalent interactions between the substrate and the enzyme's active site determines the binding affinity and specificity.
- Co-factors and co-enzymes: Many enzymes require co-factors (metal ions) or co-enzymes (organic molecules) for their activity. These molecules can directly participate in substrate binding or catalysis, enhancing specificity.
- Allosteric regulation: Allosteric effectors can bind to sites other than the active site, influencing the enzyme's conformation and thus its substrate affinity and catalytic activity. This mechanism contributes significantly to the regulation of enzyme specificity in response to cellular signals.
- Post-translational modifications: Modifications such as glycosylation, phosphorylation, or ubiquitination can alter the enzyme's conformation, influencing substrate binding and specificity.
Examples of Enzyme Specificity
Many enzymes exhibit high levels of specificity, catalyzing only a single reaction or a limited set of reactions with closely related substrates. For example:
- Hexokinase phosphorylates various hexoses, demonstrating broad specificity within a limited substrate class.
- Sucrase specifically hydrolyzes sucrose, a disaccharide, highlighting a narrower substrate specificity.
- Trypsin cleaves peptide bonds specifically on the carboxyl side of lysine and arginine residues, indicating high specificity for certain amino acid residues.
- RNA polymerase exhibits extreme specificity, recognizing and binding only to specific DNA sequences, ensuring accurate gene transcription.
These examples highlight the diversity of enzyme specificity, ranging from broad to extremely narrow substrate preferences. The specific level of specificity is determined by the enzyme's precise three-dimensional structure and the nature of its interactions with substrates.
Conclusion
Enzyme specificity is a critical aspect of cellular function, ensuring that metabolic pathways proceed efficiently and accurately. This specificity is a direct outcome of the precisely defined three-dimensional structure of the enzyme, determined by its primary sequence and subsequent folding into secondary, tertiary, and sometimes quaternary structures. The active site, a crucial region shaped by the enzyme's structure, plays a central role in recognizing and binding specific substrates through numerous non-covalent interactions. While the lock-and-key model provides a simplified representation, the induced-fit model more accurately reflects the dynamic interplay between enzyme and substrate, emphasizing the conformational changes that enhance binding and catalysis. A deep understanding of the relationship between protein structure and enzyme specificity is essential for advancing our knowledge of biological processes and developing novel therapeutic interventions. Future research in this area will undoubtedly uncover further complexities and nuances in the intricate dance between enzyme and substrate, furthering our understanding of life itself.
Latest Posts
Latest Posts
-
How Many Calories Are In 1g Of Uranium
Mar 19, 2025
-
10 Of 100 Is How Much
Mar 19, 2025
-
How Many Meters Is 150 Ft
Mar 19, 2025
-
105 Out Of 150 As A Percentage
Mar 19, 2025
-
Why Are The Noble Gases The Least Reactive Elements
Mar 19, 2025
Related Post
Thank you for visiting our website which covers about Explain How Protein Structure Is Involved In Enzyme Specificity . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
