Trna Uses What To Match To The Mrna
Kalali
Apr 01, 2025 · 6 min read
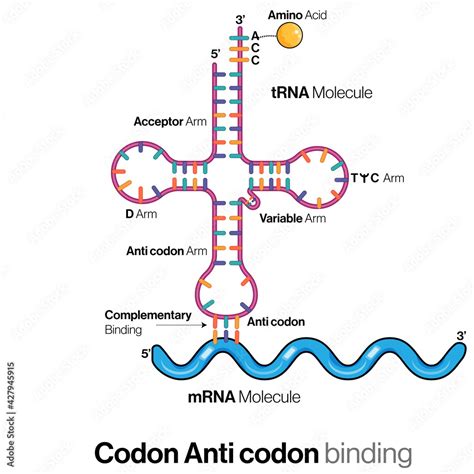
Table of Contents
tRNA Uses Anticodons to Match to mRNA: A Deep Dive into Translation
Translation, the process of protein synthesis, is a fundamental pillar of molecular biology. At the heart of this intricate mechanism lies the interaction between transfer RNA (tRNA) and messenger RNA (mRNA). This article will explore the crucial role of tRNA in decoding mRNA, focusing on how tRNA uses its anticodon to precisely match with the mRNA's codon, ensuring the accurate assembly of amino acids into polypeptide chains. We will delve into the structure of tRNA, the wobble hypothesis, aminoacyl-tRNA synthetases, and the implications of errors in this crucial matching process.
Understanding the Players: tRNA and mRNA
Before diving into the specifics of the match, let's revisit the roles of tRNA and mRNA in translation.
Messenger RNA (mRNA): The Blueprint
mRNA acts as the intermediary molecule carrying the genetic information from DNA to the ribosomes, the protein synthesis machinery. The genetic code is encoded within mRNA as a sequence of codons, each codon consisting of three consecutive nucleotides (e.g., AUG, UUU, GCA). Each codon specifies a particular amino acid, or signals the start or stop of protein synthesis. The sequence of codons in mRNA dictates the amino acid sequence of the resulting protein.
Transfer RNA (tRNA): The Adapter Molecule
tRNA plays the vital role of a molecular adapter, bridging the gap between the nucleic acid language of mRNA and the amino acid language of proteins. Each tRNA molecule carries a specific amino acid attached to its 3' end. Crucially, tRNA also possesses a unique sequence of three nucleotides called the anticodon, which is complementary to a specific mRNA codon. The anticodon enables tRNA to recognize and bind to its corresponding codon on the mRNA molecule.
The Anticodon: The Key to Accurate Matching
The anticodon is the key to the specificity of tRNA-mRNA interaction. Through complementary base pairing (A with U, and G with C), the anticodon on the tRNA precisely recognizes and binds to the corresponding codon on the mRNA. This ensures that the correct amino acid is added to the growing polypeptide chain during translation.
Complementary Base Pairing: A Closer Look
The interaction between the anticodon and the codon relies on the principle of complementary base pairing:
- Adenine (A) on the anticodon pairs with Uracil (U) on the codon.
- Guanine (G) on the anticodon pairs with Cytosine (C) on the codon.
This precise pairing is essential for accurate translation. Any mismatch can lead to the incorporation of the wrong amino acid, potentially resulting in a non-functional or even harmful protein.
The Wobble Hypothesis: Relaxing the Strict Pairing Rules
While generally strict complementary base pairing is observed, the wobble hypothesis explains how some tRNA molecules can recognize multiple codons. This flexibility is achieved through less stringent base pairing at the third position (3') of the codon. The wobble position allows a single tRNA anticodon to interact with more than one codon specifying the same amino acid, thus reducing the number of tRNAs needed for translation. This is crucial for efficiency, as it minimizes the number of different tRNA molecules required by the cell.
Wobble Base Pairing Examples:
The wobble position allows for non-Watson-Crick base pairing. For instance:
- Inosine (I), a modified base found in some anticodons, can pair with U, C, or A.
- U in the anticodon can pair with A or G in the codon.
This wobble phenomenon contributes significantly to the efficiency and robustness of the translation process.
Aminoacyl-tRNA Synthetases: Charging the tRNA
Before tRNA can participate in translation, it needs to be "charged" with its specific amino acid. This crucial step is catalyzed by aminoacyl-tRNA synthetases, a family of enzymes, one for each amino acid. These enzymes recognize both the specific tRNA and the corresponding amino acid, ensuring that the correct amino acid is attached to the correct tRNA molecule. The amino acid is attached to the 3' end of the tRNA molecule, forming an aminoacyl-tRNA complex, ready to participate in protein synthesis.
The Specificity of Aminoacyl-tRNA Synthetases:
The accuracy of aminoacyl-tRNA synthetases is paramount. Errors in aminoacylation can lead to the incorporation of incorrect amino acids into proteins, potentially compromising protein function. The high fidelity of these enzymes is essential for the overall accuracy of protein synthesis.
The Ribosome: The Protein Synthesis Factory
The ribosome serves as the protein synthesis machinery, providing a platform for the interaction between mRNA, tRNA, and other factors involved in translation. The ribosome has three binding sites for tRNA:
- A (aminoacyl) site: This site binds to the incoming aminoacyl-tRNA complex.
- P (peptidyl) site: This site holds the tRNA carrying the growing polypeptide chain.
- E (exit) site: This site releases the uncharged tRNA after it has donated its amino acid.
The ribosome facilitates the precise pairing of the tRNA anticodon with the mRNA codon, ensuring the sequential addition of amino acids to the growing polypeptide chain.
Errors in tRNA-mRNA Matching: Consequences and Mechanisms of Correction
While the accuracy of tRNA-mRNA matching is remarkably high, errors can occasionally occur. These errors can arise from various sources, including:
- Errors in aminoacylation: Incorrect amino acid attachment to tRNA.
- Mispairing of anticodon and codon: Wobble pairing can lead to occasional mismatches.
- Errors in ribosome function: Incorrect positioning of tRNA on the ribosome.
The consequences of such errors can be severe, ranging from minor functional defects to the production of non-functional or even toxic proteins. However, cells have evolved mechanisms to minimize these errors, including proofreading functions of aminoacyl-tRNA synthetases and the ribosome itself. Additionally, cellular quality control mechanisms, such as protein degradation pathways, help to eliminate misfolded or non-functional proteins resulting from translation errors.
tRNA Structure: A Closer Examination
The three-dimensional structure of tRNA is crucial for its function. The molecule folds into a characteristic cloverleaf structure, stabilized by hydrogen bonds between complementary base pairs. This cloverleaf structure further folds into an L-shaped conformation, bringing the anticodon loop and the amino acid attachment site into close proximity. This specific conformation is essential for the interaction with the ribosome and mRNA.
Conclusion: The Precision of tRNA-mRNA Matching
The precise matching of tRNA anticodons to mRNA codons is essential for accurate protein synthesis. The sophisticated interplay between tRNA structure, aminoacyl-tRNA synthetases, the wobble hypothesis, and the ribosome ensures a remarkably high level of accuracy in translation. While errors can occur, cellular mechanisms are in place to minimize their impact, maintaining the integrity of the proteome. Further research continues to unravel the intricate details of this fundamental biological process, shedding light on its regulation and implications in various biological contexts, including disease and evolution. The fidelity of tRNA-mRNA matching underscores the elegance and efficiency of life's fundamental processes.
Latest Posts
Latest Posts
-
How Many Cups In 5 Quarts Of Water
Apr 02, 2025
-
12 Out Of 18 As A Percentage
Apr 02, 2025
-
What Is A 36 Out Of 50
Apr 02, 2025
-
Describe The Cross Section Of The Rectangular Prism
Apr 02, 2025
-
7 Liters Is How Many Gallons
Apr 02, 2025
Related Post
Thank you for visiting our website which covers about Trna Uses What To Match To The Mrna . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
